SQL Query to Update a Column based on Count of repeated Items in another Column
Scenario:
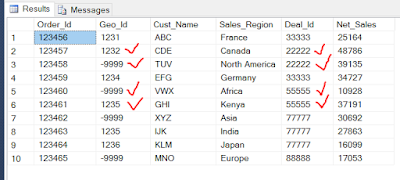
Suppose we have a table as follows
SELECT [Order_Id]
,[Geo_Id]
,[Cust_Name]
,[Sales_Region]
,[Deal_Id]
,[Net_Sales]
FROM [dbo].[tbl_Sample]
GO
In this Table , we have to update the Column Geo_Id based on the following Conditions :
A). If we have only 2 Geo_Ids, one =-9999 and Other one <> -9999 ( Eg: -9999 , 1232 ) under the same Deal_Id ( Eg : 22222) Then we need to update the Geo_Id "-9999" with another Geo_Id , say 1232.
B). If we have only one Geo_Id , as -9999 under a Deal_Id ( Eg: 88888) then we should not perform any update.
C). If we have more than two Geo_Ids , one =-9999 and Others<> -9999 (Eg : -9999,1235 1236 ) under the same Deal_Id ( Eg : 77777) then we should not perform any update.
Now we can achieve this Scenario using the below Methods :
1) Sub Query Method :
Update [tbl_Sample]
Set Geo_Id =S1.Geo_ID
From (
SELECT A.Order_Id,A.Geo_Id,A.Cust_Name,A.Sales_Region,B.Deal_Id, A.Net_Sales,B.GeoCountByDeal
FROM [tbl_Sample] A
Inner Join (
SELECT COUNT(Distinct Geo_Id) as GeoCountByDeal, B.Deal_Id
FROM [tbl_Sample] B
GROUP BY B.Deal_Id
) AS B
ON A.Deal_Id = B.Deal_Id
WHERE GeoCountByDeal=2 and A.Geo_Id<>'-9999'
-- ORDER by A.Deal_Id
) S1
Where ( [tbl_Sample].Deal_Id=S1.Deal_Id and [tbl_Sample].Geo_Id=-9999 and S1.GeoCountByDeal=2 )
Select * From [tbl_Sample]
GO
2) Common Table Expressions (CTEs) Method :
WITH MyCTE
AS
(
SELECT A.Order_Id,A.Geo_Id,A.Cust_Name,A.Sales_Region,B.Deal_Id, A.Net_Sales,B.GeoCountByDeal
FROM [tbl_Sample] A
Inner Join (
SELECT COUNT(Distinct Geo_Id) as GeoCountByDeal, B.Deal_Id
FROM [tbl_Sample] B
GROUP BY B.Deal_Id
) AS B
ON A.Deal_Id = B.Deal_Id
WHERE GeoCountByDeal=2 and A.Geo_Id<>'-9999'
-- ORDER by A.Deal_Id
)
Update [tbl_Sample]
Set [tbl_Sample].Geo_Id =MyCTE.Geo_ID
From MyCTE
Where ( [tbl_Sample].Deal_Id=MyCTE.Deal_Id and [tbl_Sample].Geo_Id=-9999 and MyCTE.GeoCountByDeal=2 )
Select * From [tbl_Sample]
;
GO
Output after Update:
--------------------------------------------------------------------------------------------------------
Thanks, TAMATAM ; Business Intelligence & Analytics Professional
--------------------------------------------------------------------------------------------------------
Scenario:
Suppose we have a table as follows
SELECT [Order_Id]
,[Geo_Id]
,[Cust_Name]
,[Sales_Region]
,[Deal_Id]
,[Net_Sales]
FROM [dbo].[tbl_Sample]
GO
In this Table , we have to update the Column Geo_Id based on the following Conditions :
A). If we have only 2 Geo_Ids, one =-9999 and Other one <> -9999 ( Eg: -9999 , 1232 ) under the same Deal_Id ( Eg : 22222) Then we need to update the Geo_Id "-9999" with another Geo_Id , say 1232.
B). If we have only one Geo_Id , as -9999 under a Deal_Id ( Eg: 88888) then we should not perform any update.
C). If we have more than two Geo_Ids , one =-9999 and Others<> -9999 (Eg : -9999,1235 1236 ) under the same Deal_Id ( Eg : 77777) then we should not perform any update.
Now we can achieve this Scenario using the below Methods :
1) Sub Query Method :
Update [tbl_Sample]
Set Geo_Id =S1.Geo_ID
From (
SELECT A.Order_Id,A.Geo_Id,A.Cust_Name,A.Sales_Region,B.Deal_Id, A.Net_Sales,B.GeoCountByDeal
FROM [tbl_Sample] A
Inner Join (
SELECT COUNT(Distinct Geo_Id) as GeoCountByDeal, B.Deal_Id
FROM [tbl_Sample] B
GROUP BY B.Deal_Id
) AS B
ON A.Deal_Id = B.Deal_Id
WHERE GeoCountByDeal=2 and A.Geo_Id<>'-9999'
-- ORDER by A.Deal_Id
) S1
Where ( [tbl_Sample].Deal_Id=S1.Deal_Id and [tbl_Sample].Geo_Id=-9999 and S1.GeoCountByDeal=2 )
Select * From [tbl_Sample]
GO
2) Common Table Expressions (CTEs) Method :
WITH MyCTE
AS
(
SELECT A.Order_Id,A.Geo_Id,A.Cust_Name,A.Sales_Region,B.Deal_Id, A.Net_Sales,B.GeoCountByDeal
FROM [tbl_Sample] A
Inner Join (
SELECT COUNT(Distinct Geo_Id) as GeoCountByDeal, B.Deal_Id
FROM [tbl_Sample] B
GROUP BY B.Deal_Id
) AS B
ON A.Deal_Id = B.Deal_Id
WHERE GeoCountByDeal=2 and A.Geo_Id<>'-9999'
-- ORDER by A.Deal_Id
)
Update [tbl_Sample]
Set [tbl_Sample].Geo_Id =MyCTE.Geo_ID
From MyCTE
Where ( [tbl_Sample].Deal_Id=MyCTE.Deal_Id and [tbl_Sample].Geo_Id=-9999 and MyCTE.GeoCountByDeal=2 )
Select * From [tbl_Sample]
;
GO
Output after Update:
--------------------------------------------------------------------------------------------------------
Thanks, TAMATAM ; Business Intelligence & Analytics Professional
--------------------------------------------------------------------------------------------------------